Use statismo models for elastic mesh registration in R
14 Aug 2014I finally found some time to properly implement statismo models into the surface matching functions AmbergRegister and gaussMatch from my package mesheR.
Here is an example how to do this:
CAVEAT:
You need to install latest version from master branch of mesheR, RvtkStatismo and Morpho !!require(RvtkStatismo)
require(Rvcg)
require(Morpho)
require(mesheR)
require(rgl)
data(humface)
data(dummyhead)
### first create a model based on the reference
mymod <- statismoModelFromRepresenter(dummyhead.mesh,kernel = list(c(50,50),c(20,20),c(10,20),c(5,20)),ncomp = 100)## combine some Gaussian kernels
##now create an object of class BayesDeform
Bayes <- createBayes(mymod,sdmax=c(rep(3,5),ptValueNoise = 2))# this means that the first 5 elastic iterations are restricted to be within 3 standard deviations of our model and we provide some point noise for the landmarks
## setup parameters for AmbergRegister
params <- list(iterations=10)
params <- append(params, list(
# then let it increase from 0.2 to 0.6
lambda=seq(from = 0.2,to=0.6,length.out = params$iterations),
# treat \code{k} similar as \code{lambda}
k=seq(from = 1,to=params$iterations,by=1),
useiter=FALSE # iteratively deform dummyhead onto humface
))
## setup parameters for some additional rigid wiggling (ICP)
rigid <- list(iterations=60,subsample=200,rhotol=pi/2,uprange=0.3)#here we specify an overlap between reference and target of 30%
## run the matching
map <- AmbergRegister(dummyhead.mesh, humface, lm1=dummyhead.lm, lm2=humface.lm, iterations=params$iterations,k=params$k, lambda=params$lambda, useiter=params$useiter,rigid=rigid,Bayes=Bayes)
## show the results
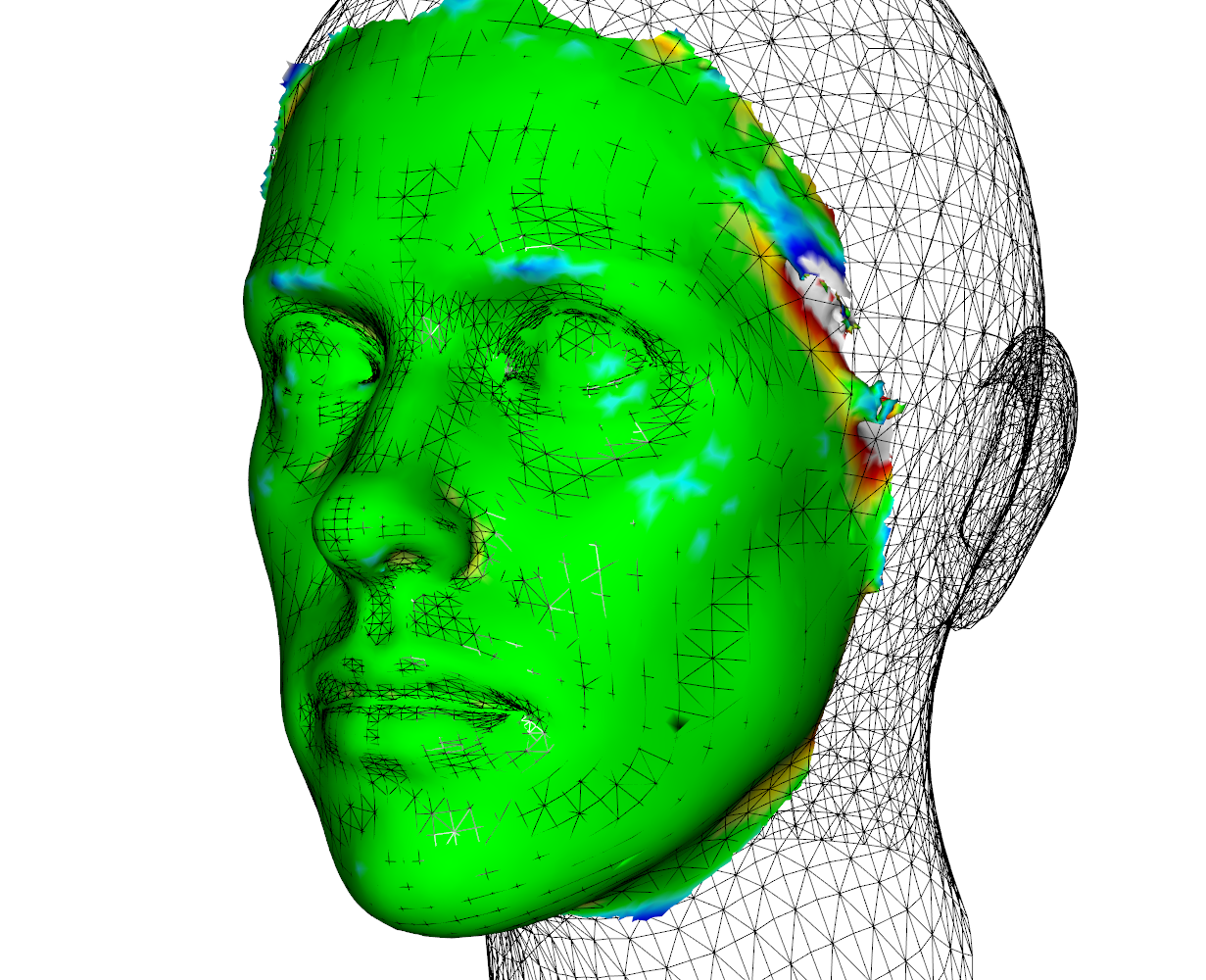
meshDist(humface,map$mesh,tol=0.5,from=-3,to=3)## create heatmap with distances (figure 1)
wire3d(map$mesh)##this is our matched mesh # figure 2
## check if landmarks are in a reasonable position
spheres3d(map$lm1,col=2)##mapped landmarks
spheres3d(humface.lm)##original landmarks
## Additionally, we can look at that shape within the model space that is closest to our surface (figure 3)
modelsurf <- PredictSample(mymod,map$mesh,T)
wire3d(modelsurf,col=2)
## we can see that the shape of the cheeks is not represented well by the model but the overall shape is


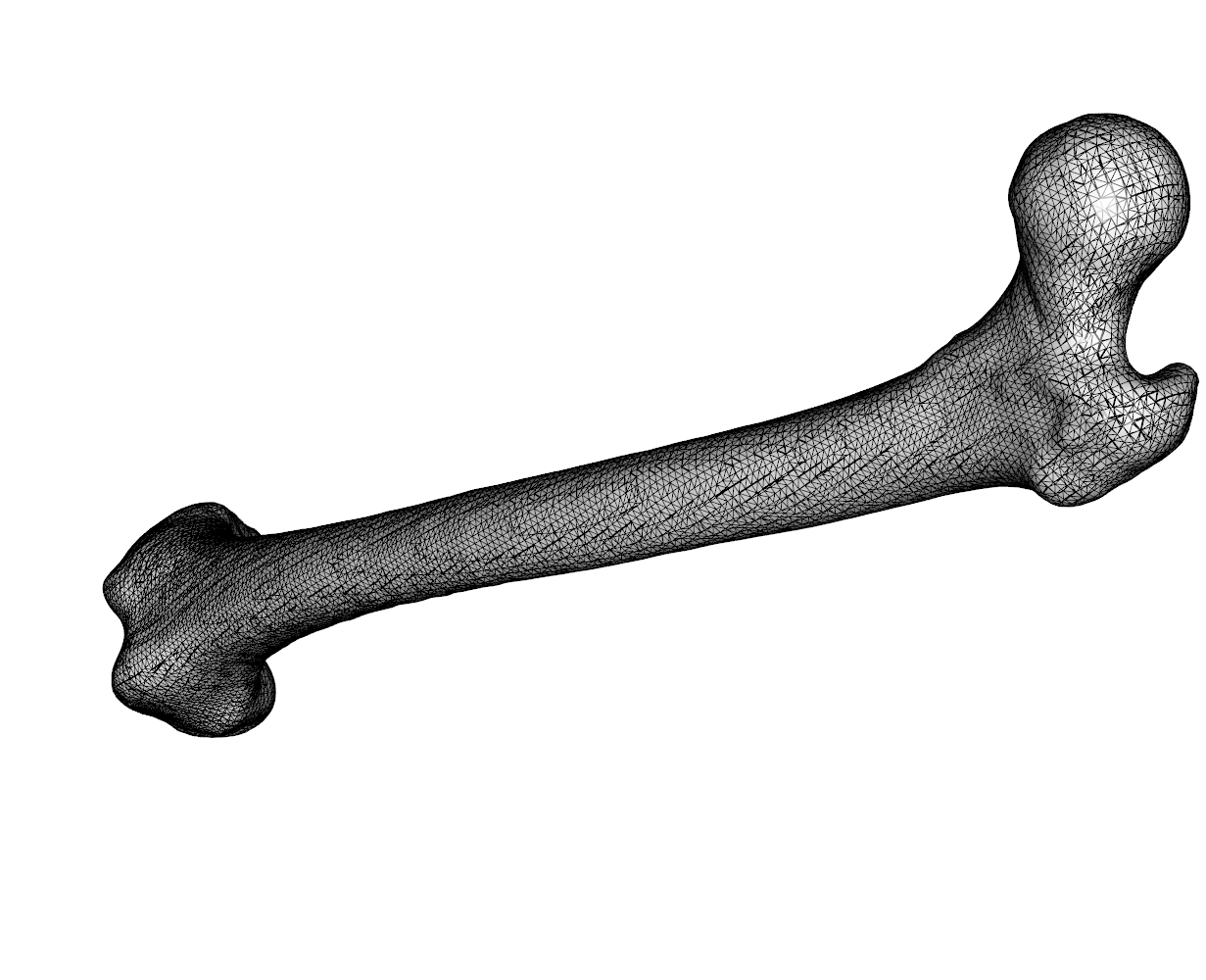
And here is an example with Marcel’s Femur surfaces:
require(RvtkStatismo)
require(Rvcg)
require(Morpho)
require(mesheR)
require(rgl)
download.file(url="https://github.com/marcelluethi/statismo-shaperegistration/raw/master/data/VSD001_femur.vtk","./VSD001_femur.vtk",method = "w")
download.file(url="https://github.com/marcelluethi/statismo-shaperegistration/raw/master/data/VSD002_femur.vtk","./VSD002_femur.vtk",method = "w")
download.file(url="https://github.com/marcelluethi/statismo-shaperegistration/raw/master/data/VSD001-lm.csv","./VSD001-lm.csv",method = "w")
download.file(url="https://github.com/marcelluethi/statismo-shaperegistration/raw/master/data/VSD002-lm.csv","./VSD002-lm.csv",method = "w")
ref <- read.vtk("VSD001_femur.vtk")
tar <- read.vtk("VSD002_femur.vtk")
ref.lm <- as.matrix(read.csv("VSD001-lm.csv",row.names=1))
tar.lm <- as.matrix(read.csv("VSD002-lm.csv",row.names=1))
mymod <- statismoModelFromRepresenter(ref,kernel=list(c(50,50),c(10,10)),ncomp = 100)
Bayes <- createBayes(mymod,sdmax = rep(6,10))##restrict first 10 iterations to model
## setup similarity and affine icps
similarity = list(iterations=10,rhotol=pi/2)
affine = list(iterations=10,rhotol=pi/2)
matchGP <- gaussMatch(ref,tar,lm1 = ref.lm,lm2=tar.lm,sigma = 30,gamma=4,smooth=1,smoothit = 10,smoothtype = "t",iterations = 15,toldist = 50,angtol = pi/2,Bayes=Bayes,similarity = similarity,affine = affine)
## view displacement field (figure below):
require(Morpho)
deformGrid3d(matchGP,ref,size=0.1,type="p")

EDIT:
I just realized, that we actually do not need smoothing (to prevent mesh-folding) if we use statistical models because the model carries us pretty close to a valid target shape:
matchGPNoSmooth <- gaussMatch(ref,tar,lm1 = ref.lm,lm2=tar.lm,sigma = 30,gamma=4,smooth=NULL,iterations = 15,toldist = 50,angtol = pi/2,Bayes=Bayes,similarity = similarity,affine = affine)
Now, when looking at the deformed reference, we can still see the mesh structure produced by marching cubes (so I assume):