RANTs: How to compute an initial landmark transform to pass to ANTsR
23 Oct 2015In most cases, the initial alignment of (3D) images is problematic and having some landmarks can aid greatly in finding a suitable initial transform. After, once again, struggling with image and point spaces, I came up with a function (RANTs::landmarkTransform <- click me), that allows to calculate an initial transform from landmarks (rigid, similarity or affine and save that in ITK’s mat format.
When running ANTsR subsequently, we can then provide this transform.
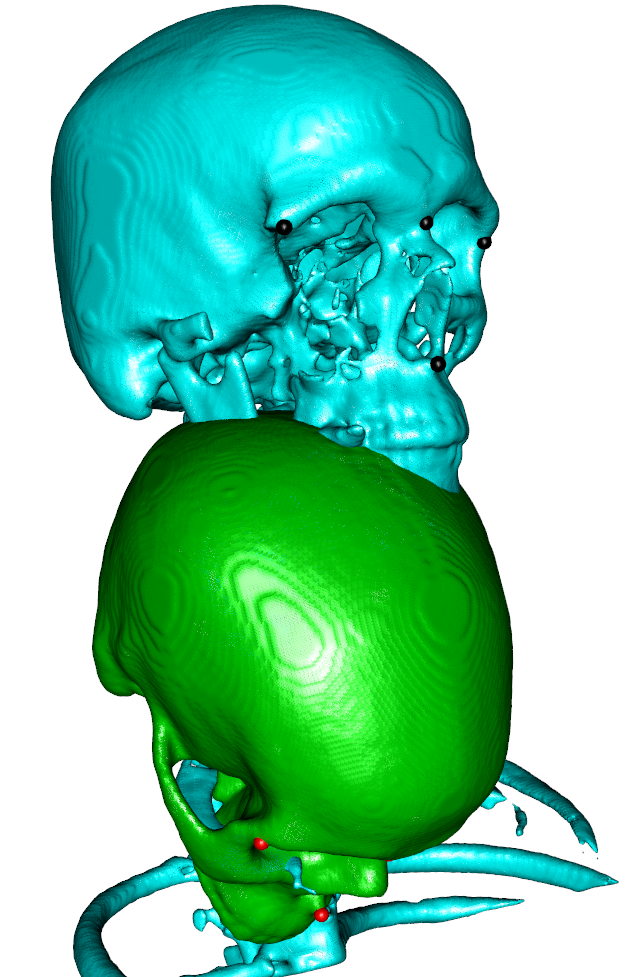
Here is an example (two binary images with 4 landmarks on each placed with Slicer) of an ANTs rigid registration preceded by an initial landmark transform. All automatic intializations failed because the regions of the images differ greatly (the moving image is only the skull while the fix image is skull + upper part of the rip cage).
We will visualize ( below ) the isosurface generated by a wrapper of Rvcg::vcgIsosurface that automatically gets origin, spacing etc. from ANTsR and SimpleITK image classes.
Example
require(RANTs)
require(RvtkStatismo)
require(rgl)
require(Rvcg)
movingimagename <- "movingimage.nii.gz"
fiximagename <- "fiximage.nii.gz"
fiximage <- antsImageRead(fiximagename)
movingimage <- antsImageRead(movingimagename)
fixLM <- read.fcsv("fixLM.fcsv")
movingLM <- read.fcsv("movingLM.fcsv")
rigidInit <- landmarkTransform(fixLM,movingLM,type = "rigid")
## show initial state and landmarks
wire3d(vcgSmooth(isosurface(movingimage,threshold=1),type="HC",iteration=4),col=3)
wire3d(vcgSmooth(isosurface(fiximage,threshold=1),type="HC",iteration=4),col=5)
spheres3d(fixLM,col=1,radius = 3)
spheres3d(movingLM,col=2,radius = 3)
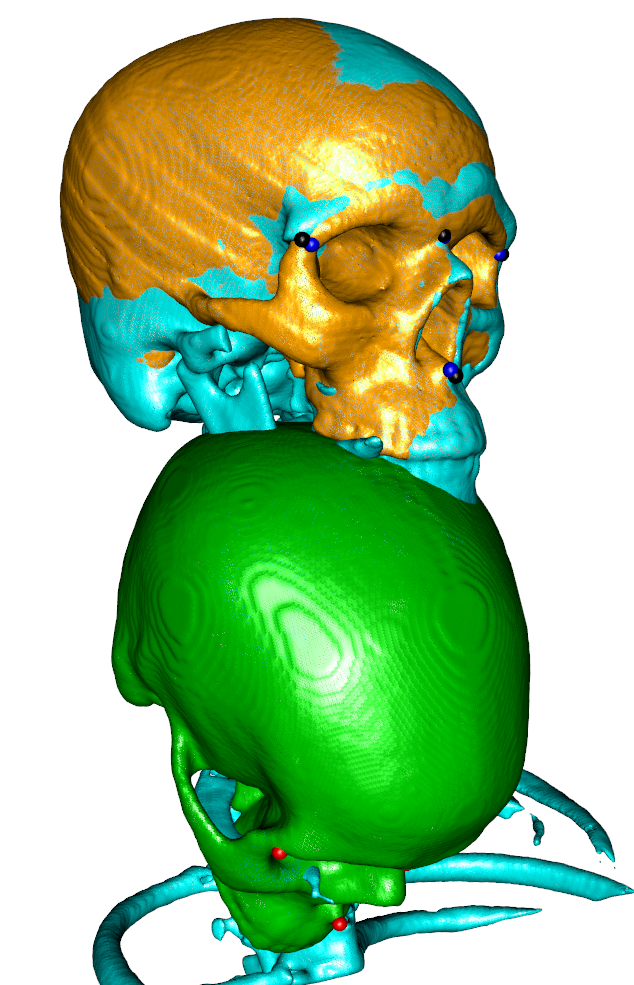
## now apply the landmark transform to the moving image and add it to the scene
movingTransformed <- antsApplyTransforms(fiximage,movingimage,rigidInit)
## apply the transform to the landmarks and note that we need to use the inverse here
landmarkTransformed <- antsTransformPoints(movingLM,rigidInit,whichtoinvert = T)
wire3d(vcgSmooth(isosurface(movingTransformed,threshold=1),type="HC",iteration=4),col="orange")
spheres3d(landmarkTransformed,col=4,radius = 3)
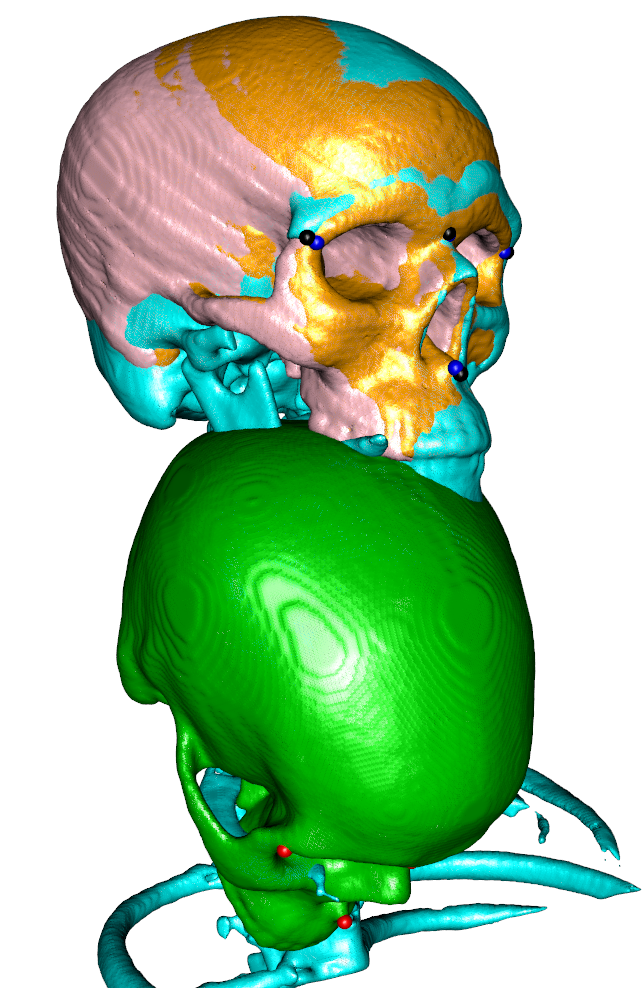
## we can also use the transform to initialize a rigid antsRegistration
myargs <- createAntsArgs(fiximagename,movingimagename,initTransform = list(rigidInit,F),affine="rigid",elastic=NULL)
antsRegistration(myargs$antsargs)
### now apply the transform and visualize the isosurface
movingRegistered <- antsApplyTransforms(fiximage,movingimage,myargs$transforms$affine)
wire3d(vcgSmooth(isosurface(movingRegistered,threshold=1),type="HC",iteration=4),col="pink")